DECREASED BREADTH OF THE ANTIBODY RESPONSE TO THE SPIKE PROTEIN OF SARS-CoV-2 AFTER VACCINATION
Lydia Horndler, Pilar Delgado, Salvador Romero-Pinedo, Marina Quesada, Ivaylo Balabanov, Rocío Laguna-Goya, Patricia Almendro-Vázquez, Miguel A. Llamas, Manuel Fresno, Estela Paz-Artal, Hisse M. van Santen, Stela Álvarez, Asunción Olmo, View ORCID ProfileBalbino Alarcón
doi: https://doi.org/10.1101/2021.08.12.21261952
This article is a preprint and has not been certified by peer review [what does this mean?]. It reports new medical research that has yet to be evaluated and so should not be used to guide clinical practice.
Abstract
The rapid development of vaccines to prevent infection by SARS-CoV-2 virus causing COVID-19 makes necessary to compare the capacity of the different vaccines in terms of development of a protective humoral response. Here, we have used a highly sensitive and reliable flow cytometry method to measure the titers of antibodies of the IgG1 isotype in blood of volunteers after receiving one or two doses of the vaccines being administered in Spain. We took advantage of the multiplexed capacity of the method to measure simultaneously the reactivity of antibodies with the S protein of the original strain Wuhan-1 and the variant B.1.1.7 (Alpha). We found significant differences in the titer of anti-S antibodies produced after a first dose of the vaccines ChAdOx1 nCov-19/AstraZeneca, mRNA-1273/Moderna, BNT162b2/Pfizer-BioNTech and Ad26.COV.S/Janssen. Most important, we found a relative reduction in the reactivity of the sera with the B.1.1.7 versus the Wuhan-1 variant after the second boosting immunization. These data allow to make a comparison of different vaccines in terms of anti-S antibody generation and cast doubts about the convenience of repeatedly immunizing with the same S protein sequence.
Competing Interest Statement
The authors have issued a patent application owned by CSIC
Funding Statement
No external funding has been received by the institution
Author Declarations
I confirm all relevant ethical guidelines have been followed, and any necessary IRB and/or ethics committee approvals have been obtained.
Yes
The details of the IRB/oversight body that provided approval or exemption for the research described are given below:
-Autonoma University Research Ethics Committee (no. #2352). -Hospital Princesa Research Ethics Committee (no. #4070).
All necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived.
Yes
I understand that all clinical trials and any other prospective interventional studies must be registered with an ICMJE-approved registry, such as ClinicalTrials.gov. I confirm that any such study reported in the manuscript has been registered and the trial registration ID is provided (note: if posting a prospective study registered retrospectively, please provide a statement in the trial ID field explaining why the study was not registered in advance).
Yes
I have followed all appropriate research reporting guidelines and uploaded the relevant EQUATOR Network research reporting checklist(s) and other pertinent material as supplementary files, if applicable.
Yes
ABSTRACT
The rapid development of vaccines to prevent infection by SARS-CoV-2 virus causing COVID-19 makes necessary to compare the capacity of the different vaccines in terms of development of a protective humoral response. Here, we have used a highly sensitive and reliable flow cytometry method to measure the titers of antibodies of the IgG1 isotype in blood of volunteers after receiving one or two doses of the vaccines being administered in Spain. We took advantage of the multiplexed capacity of the method to measure simultaneously the reactivity of antibodies with the S protein of the original strain Wuhan-1 and the variant B.1.1.7 (Alpha). We found significant differences in the titer of anti-S antibodies produced after a first dose of the vaccines ChAdOx1 nCov-19/AstraZeneca, mRNA-1273/Moderna, BNT162b2/Pfizer-BioNTech and Ad26.COV.S/Janssen. Most important, we found a relative reduction in the reactivity of the sera with the B.1.1.7 versus the Wuhan-1 variant after the second boosting immunization. These data allow to make a comparison of different vaccines in terms of anti-S antibody generation and cast doubts about the convenience of repeatedly immunizing with the same S protein sequence.
INTRODUCTION
Vaccines to prevent the worst effects of infections by SARS-CoV-2 virus causing COVID-19 have been rapidly developed. This unprecedented fast development of vaccines has allowed a significant reduction in the number of deaths caused by COVID-19 as well as in the number of patients admitted to the intensive care units of hospitals. In Spain, the immunity reached by the widespread vaccination, and immunity due to previous infections, is likely responsible for the low number of deaths caused by the so-called the fifth-wave by the end of July 2021, in spite reaching a number of reported infections similar to that of previous waves in the fall of 2020 and beginning of year 2021 that caused a much higher number of deaths (https://covid19.who.int/region/euro/country/es). The vaccines that have been more commonly administered in Spain are the mRNA vaccines BNT162b2 (BNT) generated by BioNTech/Pfizer, mRNA-1273 by Moderna, and the adenovirus-based vaccines ChaAdOx1 nCov-19 (ChAd) and Ad26.COV2.S (Ad26) by Oxford University/AstraZeneca and Janssen/Johnsson&Johnsson, respectively. All vaccines were demonstrated to be safe and protective in clinical trials1–4, although reports of rare cases of thrombotic thrombocytopenia associated to both the ChAd and Ad26 vaccines have been published5,6. However, the emergence of variants of concern (VOC) of SARS-CoV-2, specifically the Delta variant (B.1.617) with a very high transmission capacity7 is threatening the vaccination strategies in Europe and the USA, given the fact that the efficacy of vaccines against the Delta variant is reduced8, and fully vaccinated people can become infected and spread the virus efficiently to others9. The possibility of giving a third boosting vaccination to immunosuppressed patients to increase the titers of antibodies to the spike (S) protein is being considered10. Moreover, the spread of the B.1.617 variant even in fully vaccinated people is making authorities of different countries consider the possibility of administering a third dose to the general population (https://english.elpais.com/society/2021-07-23/everything-pointing-to-need-for-a-third-covid-19-vaccine-dose-says-spanish-health-minister.html). The capacity of the B.1.617 variant to infect vaccinated individuals seems to correlate with its ability to escape neutralization by antibodies produced in response to vaccines7.
Serological tests are usually established by detecting the presence of viral antigen-specific IgG or IgM in the serum of individuals using recombinant fragments of the S or N proteins and tests based on ELISA or lateral flow assay11,12. A disadvantage of those tests is that neutralizing antibodies are not directed against the N protein and that recombinant fragments of the S protein miss the quaternary structure of the protein trimer, which is the native form of the spike protein in the viral envelope. Therefore, part of the neutralizing antibodies directed against the native S trimer could be missed in serological tests based on the expression of recombinant proteins. Recently, we have described a flow cytometry (FC) test that makes use of the cell line of hematopoietic origin Jurkat to express the native S protein of SARS-CoV-2 together with a truncated form of the human EGF receptor (huEGFRt) as a normalizing tool 13. The method allows the accurate determination of seropositivity and the measurement of antibody titers in sera from post-convalescent patients13. Here, we have used this FC method to compare the production of antibodies against the S protein in response to the prime immunization and the booster immunization of the four most common vaccines being used in Spain. In addition, we have used the multiplexed capacity of the method to simultaneously measure the reactivity of sera from vaccinated individuals with the S protein of the original Wuhan-1 isolate versus the B.1.1.7 (Alpha) mutant as an example of VOC. Our results help to stir the debate about the convenience of a third dose of the same vaccines.
RESULTS
We previously used a lentiviral vector to express the full spike “S” protein of SARS-CoV2 with the original Wuhan-1 sequence, followed by a truncated human EGFR protein (huEGFRt) linked by a T2A self-cleaving sequence in transduced cells13. This system allows expression of the two proteins from a monocystronic mRNA. We produced transducing supernatants to express the construct in the human leukemic cell line Jurkat. Taking advantage of the fact that the expression of S protein is coupled to that of huEGFRt, we calculated a ratio of mean fluorescence intensities of antibodies against S versus an antibody against huEGFR as direct estimation of the relative quantity of immunoglobulins against the S protein in sera or other fluids from different donors. In order to detect simultaneously the presence of antibodies against the S protein of the B.1.1.7 VOC of SARS-CoV-2, we used a similar strategy, cloning the S sequence of the variant (Fig. EV1) into the same lentiviral vector but replacing the huEGFRt reporter by EGFP, and generated Jurkat cells estably expressing the S protein of the B.1.1.7 variant and EGFP. In this manner, we could mix the original Jurkat cells expressing the S protein of the Wuhan-1 strain (from now on termed Jurkat-Swuhan) with equal numbers of Jurkat cells expressing the S protein of the B.1.1.7 variant (from now on termed Jurkat-SB117) and incubate the cell mixture with dilutions of sera of patients recovered from infection with SARS-CoV-2 or vaccinated. The analysis by flow cytometry allowed to gate on the GFP+ cells to calculate the ratio of [fluorescence anti-S/GFP fluorescence] as a measurement of the titer of antibodies of the IgG1 isotype against the S protein of the B.1.1.7 variant (Fig. 1A). Simultaneously, we could gate on the EGFR+ cells to calculate the ratio of [fluorescence anti-S/fluorescence anti-EGFR] as a measurement of the titer of antibodies of the IgG1 isotype against the S protein of the Wuhan-1 variant. Figure 1A shows examples of how the plots of IgG1 anti-S and either GFP or anti-EGFR appear for a positive control serum and a negative control serum. Using this procedure, we titrated sera from donors that were diagnosed as infected either with the original Wuhan-1 or the B.1.1.7 variants. For each serum dilution, the S/GFP ratio (B.1.1.7 variant) and the S/EGFR ratio (Wuhan-1 strain) were calculated and plotted. A lineal regression curve was fitted to the data and the result is shown in Fig. 1B. The sera of donors that were infected with the Wuhan-1 strain reacted almost as intensely with the B.1.1.7 variant as with the Wuhan-1 strain, and vice versa. However, the reactivity of the sera from patients recovered of infection with B.1.1.7 was slightly better with Jurkat-SB117 than with Jurkat-Swuhan. This was reflected in the fitting lines for those sera in the S/EGFR MFI (x-axis) versus the S/GFP MFI (y-axis) plot to be slightly above those for sera from patients recovered from infection with the Wuhan-1 strain (Fig. 1B). This fact was also reflected by plotting the slopes of the fitting lines (Fig. 1C). These results suggest that most of the antibodies against the S protein generated by patients recovered from infection reacted with both strains of SARS-CoV-2. Nonetheless, there were also differences in line with the sequence divergence of both strains (Fig. EV1). These data suggested that we could make use of the reactivity of sera with Jurkat-Swuhan and Jurkat-SB117 to determine the relative capacity of the anti-S antibodies to bind to each of the two variants.
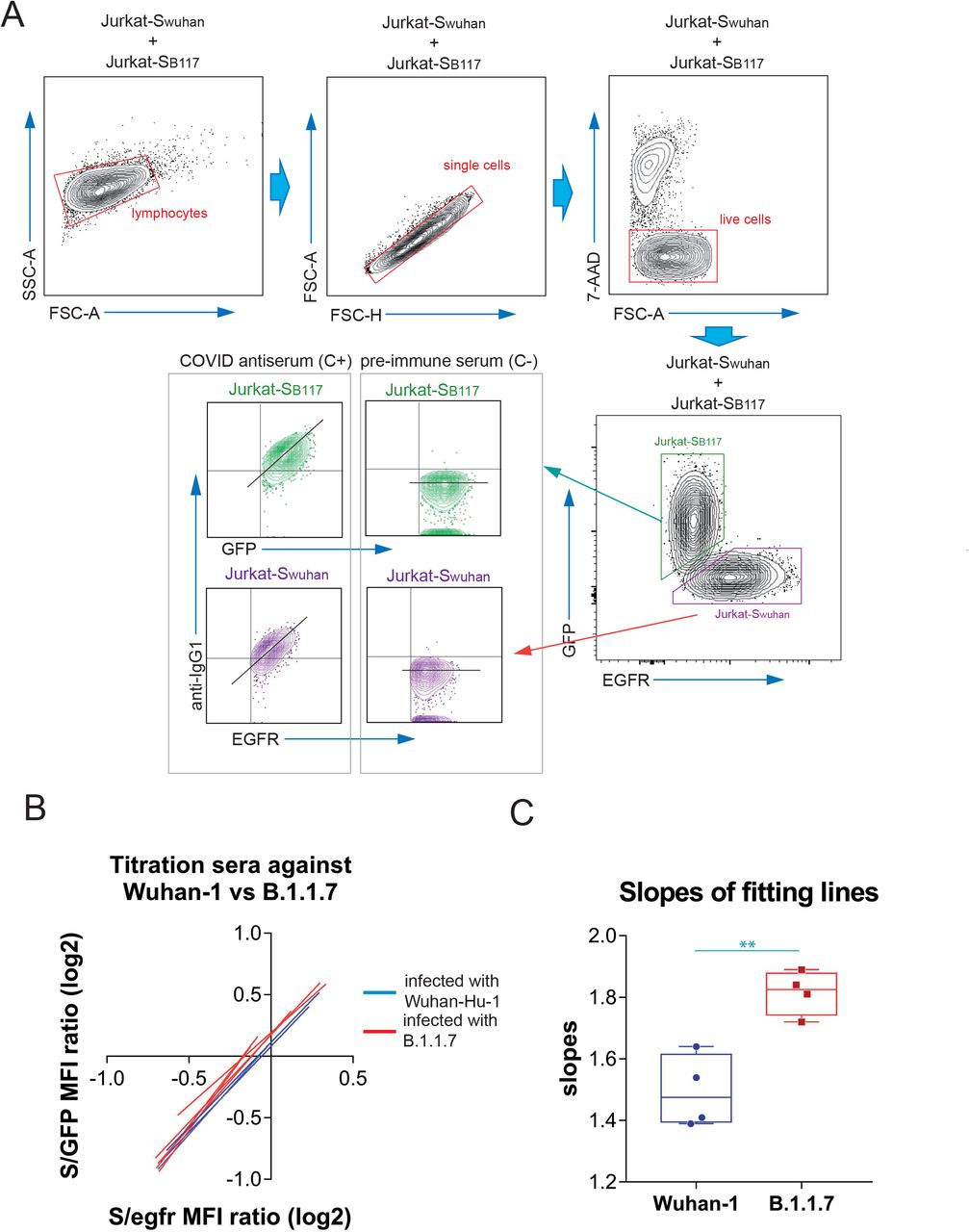
Figure 1.
Detection of antibodies in human sera binding the native S protein of the Wuhan-1 and B.1.1.7 variants of SARS-CoV-2 expressed in Jurkat-S cells.
(A) Gating strategy to measure binding to the Wuhan-1 and B.1.1.7 variants. Jurkat-Swuhan and Jurkat-SB117 cells were mixed in 1:1 ratio and incubated with either a 1:50 dilution of a positive control serum (C+) from a patient of the first wave in 2020 or a necative control serum (C-) obtained before the COVID-19 pandemics. Cells were subsequently incubated with mouse anti-human IgG1-PE and mouse anti-human EGFR-BV421 antibodies. After incubation, cells were gated according to lymphocyte FSC-A and SSC-A and subsequently on 7-AAD negative cells (7-AAD+ are dead cells). Live cells were subsequently gated on the FL1 channel (for GFP+ cells) and on the FL7 channel (huEGFRt+ cells). Both GFP+ and huEGFRt+ cells were separately analyzed for the fluorescence intensity of anti-IgG1 (S protein, FL2) versus GFP (for Jurkat-SB117) or for the fluorescence intensity of anti-IgG1 (S protein, FL2) versus EGFR (for Jurkat-Swuhan). A diagonal lane is plotted on the bicolor contour plots corresponding to the C+ staining to indicate a relationship between S protein expression and either GFP or huEGFRt.
(B) Differential binding to Jurkat-SB117 and Jurkat-Swuhan cells of four different sera tested positive by PCR for the Wuhan-1 strain (blue lines) and four different sera tested positive by PCR for the B.1.1.7 variant. 3-fold serial dilutions (from 1: 50 to 1:36450) were made for each serum and the ratios of the mean fluorescence intensities (MFI) of anti-S binding versus GFP or versus EGFR were plotted. The graph shows the fitting of all data points to straight lines. The x-axis and the y-axis show the ratio values in logarithmic scale.
(C) Box and whiskers plot of all datapoints corresponding to the slopes of the fitting lines calculated in (B). A two-tailed t test was used to assess significance. **, p<0.01.
Using this system, we decided to carry out a retrospective study to compare the generation of anti-S antibodies reactive with the Wuhan-1 and the B.1.1.7 variants in response to the most common vaccines being employed at present in Spain and compare this response with that of patients recovered from infection by SARS-CoV-2 during the first wave of the pandemics (https://covid19.who.int/region/euro/country/es) between March and May 2020 (patients 2020, Fig. 2 and Table 1) and with that of patients recovered from infection during the fourth wave of the pandemics between March and May 2021 (patients 2021, Fig. 2 and Table 1). Patients 2020 were likely infected by the Wuhan-1 strain and patients 2021 were likely to be infected by the B.1.1.7 strain, given its preponderance at that time (https://theconversation.com/the-uk-variant-is-likely-deadlier-more-infectious-and-becoming-dominant-but-the-vaccines-still-work-well-against-it-156951). We first compared sera from volunteers that received the first dose of the vaccine 3-4 weeks earlier. As shown in Fig. 2A, donors that had received the first dose of the mRNA-1273/Moderna (Moderna) and the BNT162b2/Pfizer-BioNTech (BNT) presented significantly higher titers of IgG1 anti-S antibodies of the Wuhan-1 strain than Patients 2020. However, donors that had been immunized with the ChAdOx1 nCov-19/AstraZeneca-Oxford University vaccine (ChAd) generated lower titers of antibodies against the Wuhan-1 strain than Patients 2020 and Patients 2021 and also than donors immunized with the Moderna and BNT vaccines (Fig. 2A). Although the number of samples obtained from donors immunized with the Ad26.COV2.S/Janssen-Johnsson&Johnson vaccine (Ad26) was limited, the results point out to the two vaccines based on defective adenoviruses (ChAd and Ad26) being less efficient than those based on mRNA (Moderna and BNT) (Fig. 2A). Similar results were obtained when reactivity with the S protein of the B.1.1.7 strain was analyzed (Fig. 2B).
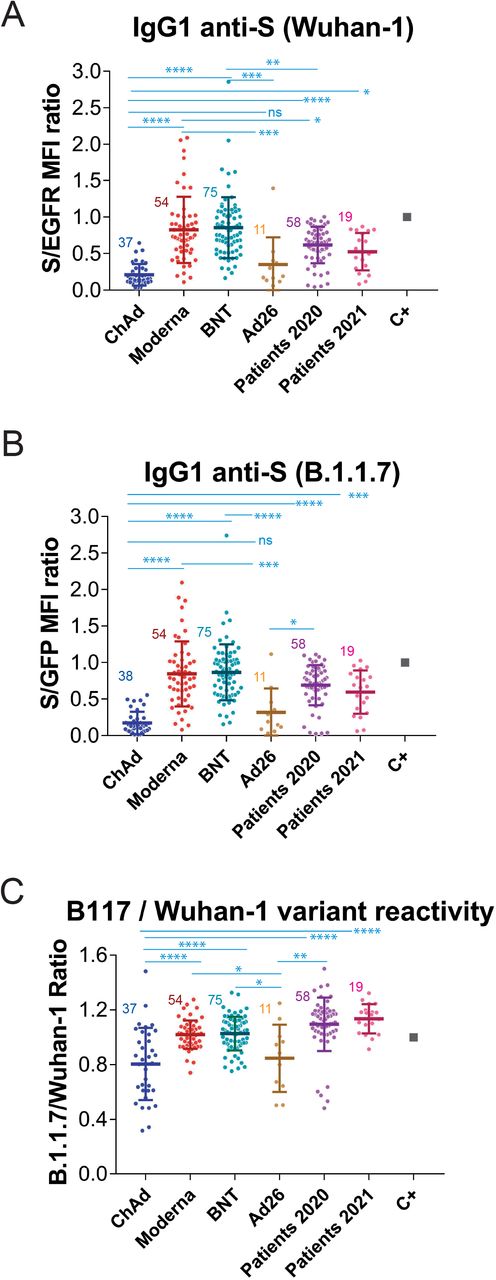
Figure 2.
Generation of anti-S protein antibodies in response to the first priming dose of different vaccines.
(A) Generation of antibodies against the S protein of the Wuhan-1 strain 3 weeks (BNT) or 4 weeks (Moderna, ChAd and Ad26) after the first, priming, dose of vaccine to previously uninfected individuals. The ratio of the mean fluorescence intensities (MFI) of anti-S staining and anti-EGFR staining was calculated for each serum sample and normalized to the value of the positive control (C+) sample. All sera were used at a 1:50 dilution. The mean±SD is shown. The number of samples for each vaccine is shown in the graph. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001; ns, not significant (one-way ANOVA test).
(B) Reactivity of antibodies from sera tested in (A) with the B.1.1.7 variant was tested simultaneously to (A).
(C) Relative reactivity of the sera tested in (A) and (B) against the B.1.1.7 and Wuhan-1 variants was measured according to the (S/GFP ratio) / (S/EGFR ratio) shown in the above panels. Statistical significance was assessed as in (A).
The combination of Jurkat-Swuhan and Jurkat-SB117 cells in the same experiment allows to calculate a ratio of ratios (S/GFP vs S/EGFR) that serves to compare sera from vaccinated volunteers in terms of their relative reactivity with the S protein of the Wuhan-1 and B.1.1.7 variants. Interestingly, the reactivity with the B.1.1.7 VOC was relatively better in individuals vaccinated with the mRNA vaccines than in those receiving adenovirus-based vaccines (Fig. 2C). These data also show that the relative reactivity of sera from Patients 2021 and even Patients 2020 with the B.1.1.7 VOC was better than that of sera from all vaccinees.
We next aimed to measure the effect of the second (booster) dose of vaccine on the generation of IgG1 anti-S antibodies against the two SARS-CoV-2 variants. Compared to the titer generated after the first, priming, dose, the second immunization with Moderna and BNT did not induce a significant increase in the reactivity with the Wuhan-1 (Fig. 3A) or B.1.1.7 (Fig. 3B) variants. By contrast, a significant increase in the recognition of both variants occurred for volunteers vaccinated with the second dose of ChAd, compared to those that had received only one dose of the same vaccine (Fig. 3A and 3B). Nevertheless, sera from volunteers who had received two doses of BNT, but not those who received two doses of Moderna, still reacted better than those from donors that had received two doses of the ChAd vaccine. Interestingly, the second dose of Moderna and the second dose of BNT caused a relative decrease in the reactivity of the antibodies with the B1.1.7 VOC in comparison with the Wuhan-1 isolate (Fig. 3C). The second dose of ChAd did not cause either an increase or a decrease in the relative recognition of the S protein of B.1.1.7 compared to Wuhan-1. However, antibodies produced in response to the second dose of either of the three vaccines were relatively less reactive with the B.1.1.7 strain than those produced by natural infection not only in 2021 but also in 2020 (Fig. 3C). Those results suggest that the booster immunization with the second dose of the three vaccines, based on the sequence of the Wuhan strain, might impoverish the reactivity of the antibodies with emergent VOC of SARS-CoV-2. The relative loss of reactivity with B.1.1.7 versus the Wuhan-1 strain after the second dose of vaccine is perhaps best seen when all titers of anti-S antibodies are plotted, independent of the vaccine used (Fig. 4A). The boosting dose caused a relative loss of reactivity with the B.1.1.7 variant, compared to the first, priming dose. In addition, sera from patients recovered after the first wave (2020) and the fourth wave (2021) presented a relative higher reactivity with the B.1.1.7 variant than those from individuals vaccinated with one or two doses (Fig. 4A).
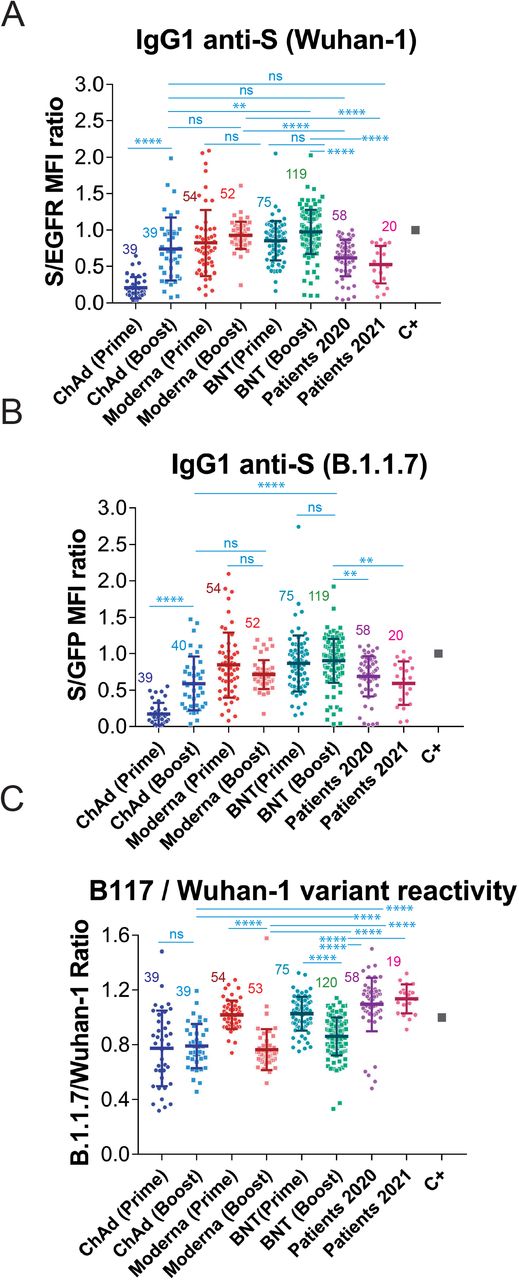
Figure 3.
Reduction in the relative recognition of the B.1.1.7 versus the Wuhan-1 variants after immunization with the second, booster, dose of vaccines.
(A&B) Generation of antibodies against the S protein of the Wuhan-1 (A) and B.1.1.7 (B) variants strain 3 weeks (BNT) or 4 weeks (Moderna, ChAd and Ad26) after the first, priming, dose of vaccine and 4 weeks (all vaccines) after the second, booster dose of vaccine. The ratio mean fluorescence intensities (MFI) of anti-S staining and anti-EGFR staining was calculated for each serum sample and normalized to the value of the positive control (C+) sample. All sera were used at a 1:50 dilution. The mean±SD is shown. The number of samples for each vaccine is shown in the graph. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001; ns, not significant (one-way ANOVA test).
(C) Relative reactivity of the sera tested in (A) and (B) against the B.1.1.7 and Wuhan-1 variants was measured according to the (S/GFP ratio) / (S/EGFR ratio) shown in the above panels. Statistical significance was assessed as in (A) and (B).
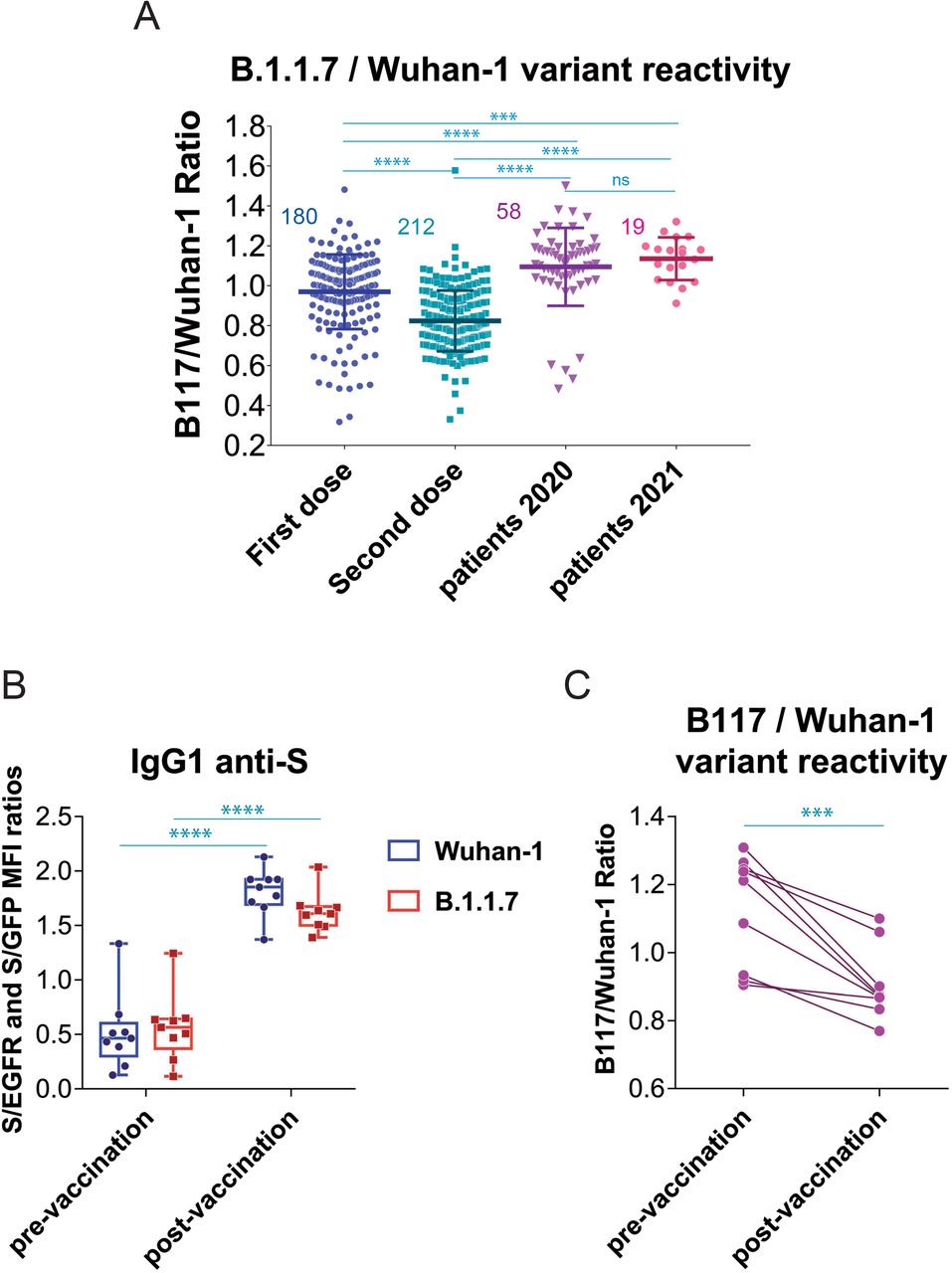
Figure 4.
Effect of vaccination on the relative recognition of the B1.1.7 and Wuhan-1 variants by sera of vaccinated individuals.
(A) Effect of the second dose of vaccine on the relative reactivity of sera with the B.1.1.7 and Wuhan-1 variants. All data of (S/GFP ratio) / (S/EGFR ratio) ratio generated after the first dose of vaccine (ChAd, Moderna, BNT and Ad26) is plotted with all data generated after the second dose of vaccine (Moderna and BNT). The mean±SD is shown. The number of samples for each vaccine is shown in the graph. *** p<0.001; **** p<0.0001; ns, not significant (one-way ANOVA test).
(B) Generation of antibodies against the S protein of the Wuhan-1 and B.1.1.7 variants in individuals (n=9) infected by SARS-CoV-2 in 2020 compared with the serum samples obtained in the same individuals 4 weeks after the first dose of BNT vaccine. Box and whiskers plot showing all points, the minimum and maximum values and the median. **** p<0.0001; (one-way ANOVA test)
(C) elative reactivity of the sera tested in (B) against the B.1.1.7 and Wuhan-1 variants measured according to the (S/GFP ratio) / (S/EGFR ratio) and plotted individually for each serum donor. *** p<0.001 (two-tailed paired t-test).
We next interrogated if the loss of relative reactivity against B.1.1.7 was also produced in patients recovered after the first wave in 2020 who received one dose of either BNT or Moderna (Table 1). Compared to the titer pre-vaccination, the priming dose of vaccine caused an increased reactivity of IgG1 antibodies against the Wuhan-1 variant and also, although at a lower extent, against the B.1.1.7 variant (Fig. 4B). Immunization of individuals who were already immune due to prior infection by SARS-CoV-2 resulted in a relative loss of reactivity with the B.1.1.7 variant, when analyzed in a per-individual base (Fig. 4C).
The effect of vaccination on the relative recognition of the S protein of the B.1.1.7 variant versus that of the Wuhan-1 strain was also followed in sera from volunteers of nursing homes who had ‘recovered after infection in 2020 and 2021 and were later vaccinated with BNT (Table 1). Serum samples were taken just before vaccination, 15 days after the first dose, 21 days after the first dose, 1 week after the second dose, and 4 weeks after the second dose of the BNT vaccine. The first dose of BNT resulted in a significant increase in antibody reactivity against the Wuhan-1 and B.1.1.7 variants 21 days after vaccination (Fig. 5A and 5B). Interestingly, and compared with samples taken at day 21 after the first priming dose, the second dose of vaccine did not cause a significant increase in serum reactivity either 1 week or 4 weeks after the booster immunization (Fig. 5A and 5B). Most important, vaccination provoked a relative loss in the capacity of recognition of the B.1.1.7 variant starting as early as 15 days after the first dose of BNT and worsening since then (Fig. 5C).
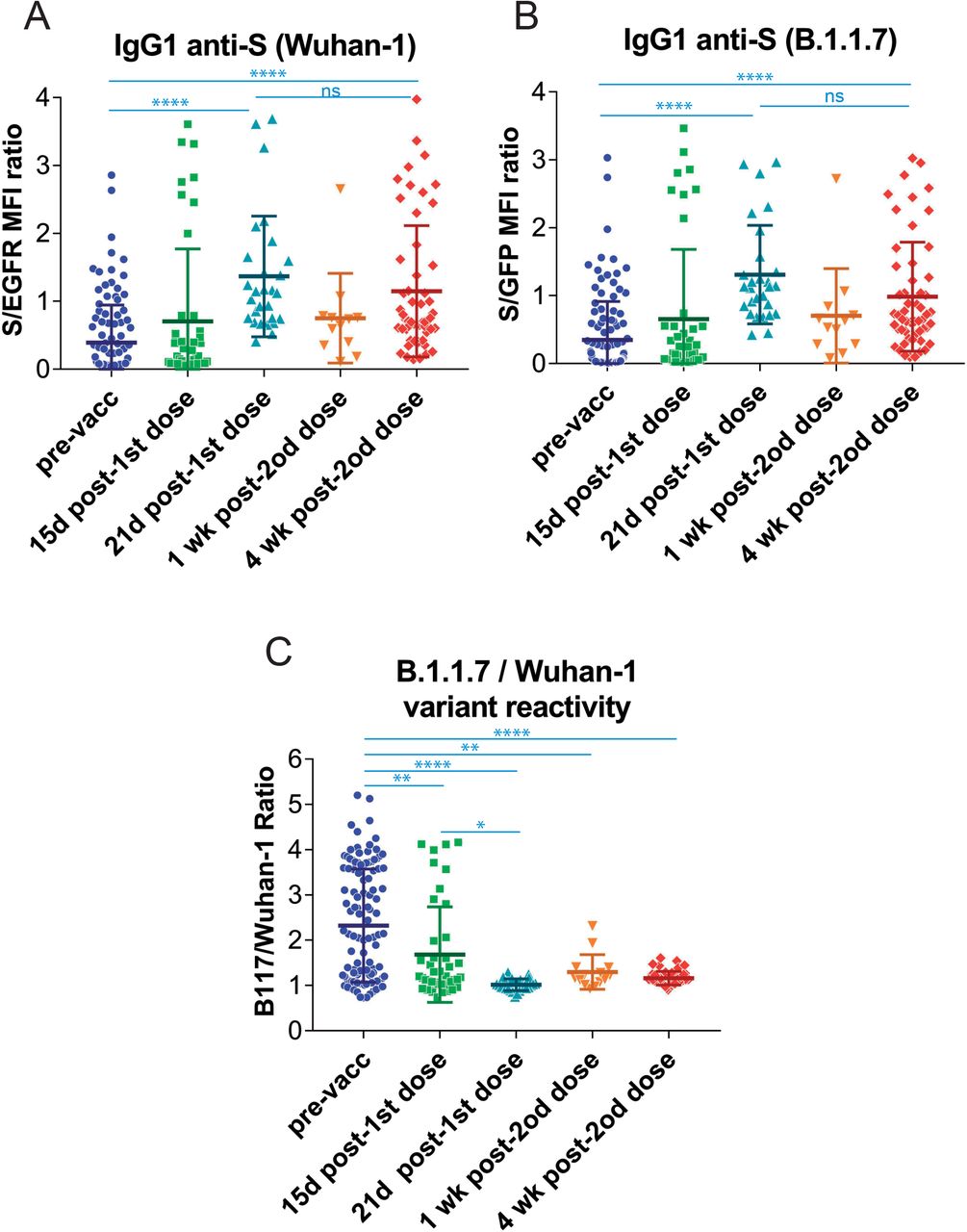
Figure 5.
Follow-up of the antibody response to vaccination of previously seropositive individuals in nursing homes.
(A&B) Generation of antibodies against the S protein of the Wuhan-1 (A) and B.1.1.7 (B) variants strain at different times before and after vaccination with the first dose and the second dose of BNT/Pfizer. The normalized S/EGFR ratio for reactivity with the Wuhan-1 strain and the S/GFP ratio for reactivity with the B.1.1.7 variant was measured in nursing home residents before the first dose (pre-vaccination, pre-vacc), 15 days and 21 days after the first dose of BNT/Pfizer and 1 week and 4 weeks after the second dose of the same vaccineAll sera were used at a 1:50 dilution. The mean±SD is shown. The number of samples for each vaccine is shown in the graph. **** p<0.0001; ns, not significant (one-way ANOVA test).
(C) Relative reactivity of the sera tested in (A) and (B) against the B.1.1.7 and Wuhan-1 variants was measured according to the (S/GFP ratio) / (S/EGFR ratio) shown in the above panels. The mean±SD is shown. * p<0.05; ** p<0.01; **** p<0.0001 (one-way ANOVA test).
DISCUSSION
In this study we have measured the production of antibodies of the IgG1 isotype to the spike protein of SARS-CoV-2 in response to the four different vaccines most frequently used in Spain. We have compared those data with antibodies present in sera from patients recovered from infection during the first wave of the COVID-19 pandemic in March-June 2020 and with sera from patients recovered from infection during the fourth wave between March-May 2021. Our results show that a priming first dose of the mRNA vaccines BNT/Pfizer and Moderna significantly induced a higher antibody response against the S protein of the Wuhan-1 strain than the two adenovirus-based vaccines ChAd and Ad26, and superior to the titers found in the cohorts of 2020 and 2021 patients. The booster immunization with a second dose did not increase the average titer of antibodies except for the ChAd vaccine. The booster immunization with ChAd resulted in antibody titers that were similar to those generated in response to the booster immunization with Moderna or to the patient cohorts of 2020 and 2021, but still lower than the titers generated in response to the booster immunization with BNT. Therefore, in summary, our results indicate that two doses of the ChAd, BNT and Moderna vaccines are able to induce antibody titers against the original Wuhan-1 strain equal or even superior to those found in post-convalescent patients. However, the message is different when we compared the relative reactivity of the antibodies against the B.1.1.7 variant versus the Wuhan-1 strain. Although antibodies made in response to vaccines based on the original Wuhan-1 strain sequence do also bind the B.1.1.7 variant, suggesting that most epitopes are conserved between both sequences, there is a relative loss of reactivity with the B.1.1.7 variant compared to the Wuhan-1 strain occurring upon administration of the booster dose of vaccine. This is somehow expected since repeated immunization with the same antigen sequence leads to the generation of higher affinity antibodies that fit better the epitopes of the immunogen. This increase in affinity has the negative side effect of reducing the “breadth” of the antibodies, that is, their capacity to bind to epitopes that differ slightly from those of the immunogen. Such effect has been observed before, for instance, as a result of vaccination with inactivated influenza virus isolated in immunization campaigns, that result in an efficient neutralization of that particular seasonal variant of influenza but results in reduced capacity to neutralize other variants14. By contrast, antibody responses to natural infection are broad and exhibit different immunodominance patterns14. Another example of the different breadth in the antibody responses elicited by natural infection versus vaccination is the long-term struggle, and still unsuccessful, to generate a vaccine that prevents infection by the tremendous diversity of clades and mutants of human immunodeficiency virus (HIV), with the general conclusion that the tested vaccines are effective to prevent infection by the strain used for immunization but not by the myriad of variants found in the field15. In regard to COVID-19, a broad and sustained polyantigenic immunoreactivity against the S protein and other viral proteins has also been found in COVID-19 patients, in this case associated to the severity of symptoms16. Our follow-up of the cohort of residents in nursing homes that were previously naturally infected with SARS-CoV-2 and later vaccinated with two doses of BNT/Pfizer shows that the first dose of vaccine increased the already existing titer of anti-S antibodies against the Wuhan-1 strain. However, the second dose did not result in further increases in the antibody titer and had the negative counterpart of reducing the relative reactivity of the antibodies with the B-1-1-7 VOC. Indeed, such loss of breadth was already detected as soon as 15 days after the first dose of vaccine, when the antibody titer against the Wuhan-1 strain had not yet increased significantly.
Although our study is limited to measuring the presence of IgG1 against the S protein of SARS-CoV-2, we think our results are relevant to evaluate the overall humoral response, given that IgG1 is the most abundant immunoglobulin in human blood and has potent effector capacities by being able to both fix complement and bind to Fc receptors17. Another limitation of our study is that we have compared the reactivity of antibodies with the Wuhan-1 strain and the B.1.1.7 (Alpha) variant and not with the Delta variant which is the most rapidly expanding VOC in Europe and the USA by the end of July 2021 (https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/fully-vaccinated-people.html). At present, we are generating a Jurkat-S cell line expressing the Delta variant to confirm that our findings on loss of antibody breadth upon vaccination for the Alpha variant also hold true for the Delta VOC. The generation of this cell line will allow to measure the reactivity of antibodies against the trimeric, native, form of the S protein, something that the serological tests based on the use of recombinant proteins do not allow.
While we are awaiting for the results with Jurkat-S expressing the Delta VOC, our results with the Alpha variant suggest that third doses of the present vaccines to the general population might not be the best approach to increase the immunity to the emerging VOCs. In our opinion, a third dose should be limited to the population that has been demonstrated to have been poorly responsive to the first two prime and boost doses.
MATERIALS AND METHODS
Cells
The human T-cell line Jurkat clone E6-1 was acquired from ATCC (TIB-152) and was maintained in complete RPMI 1640 supplemented with 5% fetal bovine serum (FBS, Sigma) in a 5% CO2 incubator. Human embryonic kidney HEK293T cells (ATCC CRL-3216) were maintained in DMEM supplemented with 10% FBS in a 5% CO2 incubator. All cell lines were routinely tested for the absence of mycoplasma. Lentiviral vector and Jurkat cell transduction
The generation of Jurkat-S cells expressing the spice protein of the Wuhan-1 (Jurkat-Swuhan) variant has been previously described13. To express the full-length spike S protein of the B.1.1.7 SARS-CoV-2 variant in Jurkat cells (Jurkat-SB117), we used the lentiviral vector based on the epHIV-7 plasmid, the spike S proteins sequences were obtained from the Public Health England database (https://www.gov.uk/coronavirus) and synthesized by the company Eurofins Genomics. The EGFP sequence was obtained from the vector pEGFP-C1 (Addgene). Plasmid construction was generated by the Gibson Assembly® method. For transduction, lentiviral-transducing supernatants were produced from transfected packaging HEK-293T cells as described previously18. Briefly, lentiviruses were obtained by co-transfecting plasmids pCMV-dR (gag/pol) and using the JetPEI transfection reagent (Polyplus Transfection). Viral supernatants were obtained after 24 and 48 hours of transfection. Polybrene (8 µg/mL) was added to the viral supernatants prior to transduction of Jurkat cells. A total of 3×105 Jurkat cells were plated in a P24 flat-bottom well in 350 µL of RPMI, and 350 µL of viral supernatant were added. Cells were centrifuged for 90 minutes at 2200 rpm and 32°C and left in culture for 24 hours. Transduced GFP+ cells were selected by FACS sorting 48 h later and expanded in culture. Human sera A total of 700 human sera were tested. A first set of 50 sera from Empireo obtained in the year 2020 after the first wave of the COVID-19 pandemics. Serum donors filled in a questionnaire to allow their clinical classification according to the following parameters: Asymptomatic, no symptoms; Mild, 3 or more of the following symptoms: non-productive cough, hyperthermia, headache, odynophagia, dyspnea, asthenia, myalgia, ageusia, anosmia, cutaneous involvement; Moderate, 3 or more of the above symptoms plus gastrointestinal symptoms, or more than 3 of the above for 7 or more days; Moderate-Severe, 3 or more of the above symptoms plus pneumonia; Severe, pneumonia requiring hospitalization and intubation. A second set of 52 serum samples were selected from the study “Immune response dynamics as predictor of COViD.19 disease evolution. Implications for therapeutic decision-making” from the Hospital Universitario La Princesa (HUP) approved by the Research Ethics Committee (no. #4070). A third set of 250 sera from nursery homes and VITRO employees. A fourth set of 40 serum samples were obtained from teachers and employees of a secondary school in Madrid (name not revealed to keep anonymity). Finally, a fifth cohort of 250 serum samples were obtained from capillary blood of volunteers working at the CBMSO in the period of January-July 2021, included in the study “ACE2 as a biomarker with utility for identification of high risk population for SARS-CoV-2 infection and prognosis of evolution in COVID-19” approved by the Research Ethics Committee (no. #2352). All participants provided written consent to participate in the study which was performed according to the EU guidelines and following the ethical principles of the Declaration of Helsinki.
Flow cytometry
A 1:1 mix of Jurkat-Swuhan and Jurkat-SB117 cells were incubated for 30 min on ice with 1:50 dilutions of human sera in phosphate-buffered saline (PBS), 1% bovine serum albumin (BSA), 0.02% sodium azide. Cells were spun for 5 min at 900 g and the pellet was resuspended in PBS-BSA buffer and spun to eliminate the excess of antibody. Two additional washes were carried out. The cell pellet was finally resuspended in a 1:200 dilution of mouse anti-human IgG1 Fc-PE (Ref.: 9054-09, Southern Biotech) and a 1:200 dilution of the Brilliant Violet 421™ anti-human EGFR Antibody (Ref.: 352911, Biolegend) in PBS-BSA. Samples were then washed, labeled with the viability dye 7AAD and analyzed on a FACSCanto II flow cytometer (Becton-Dickinson) and the data were analyzed with FlowJo software (BD). Statistics
Unpaired two-tailed Student t-tests were used to compare statistical significance between two groups of MFI values that followed a normal distribution and one-way ANOVA tests for column analysis of different groups. All data was analyzed using the GraphPad Prism 7 software. Serum samples were received coded from the providers and the experimentalists were blinded to their nature until all data analysis was finalized. Sample analysis was carried out in duplicate or triplicate and all experiments were repeated a minimum of two times.
Data Availability
All data is provided in Table 1 of the manuscript and no deposit in external repositories is required
DATA AVAILABILITY
This study includes no data deposited in external repositories. Jurkat-S cells are available for academic research upon request to B. Alarcón.
Funding
This work was funded by intramural grant CSIC-COVID19-004: 202020E081 (to B.A.) and CSIC-COVID19-004: 202020E165 (to MF). L.H has been supported by an FPI fellowship from the Spanish Ministry of Science and Innovation. I.B. has been supported by an H2020-MSCA-ITN-2016 training network grant of the European Union (GA 721358).
Author contributions
LH and PD performed research and analyzed the data; IB helped with experimentation; SR-P, MQ, RL-G, PA-V, MAL, E.-A., SA, and AO provided clinical samples and data and revised the manuscript; MF and HMvS analyzed data and supervised research, BA supervised and designed research, analyzed data and wrote the manuscript.
Conflict of interest
The authors have issued a patent application owned by CSIC.
EXPANDED VIEW FIGURES

Figure EV1.
Alingment of the amino acid sequences of the S protein of the Wuhan-1 and B.1.1.7 variants.
Amino acid mutations found in the B.1.1.7 isolate are shown in red bold type. B.1.1.7 has 5 amino acid replacements and two amino acid deletions compared to the reference Wuhan-1 strain. The sequence corresponding to the Receptor Binding Domain (RBD) is boxed. B.1.1.7 has just one amino acid (N to Y) replacement in the RBD.
Acknowledgments
We are indebted to Valentina Blanco and Tania Gómez for their expert technical assistance. We thank all volunteers of the CBM, school teachers, “Hospital 12 de Octubre” and “Hospital Universitario de la Princesa” for generously participating in the study.
REFERENCES
1.Polack, F. P. et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
2.Voysey, M. et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet Lond. Engl. 397, 99–111 (2021).
3.Baden, L. R. et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416 (2021).
4.Sadoff, J. et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N. Engl. J. Med. 384, 2187–2201 (2021).
5.Greinacher, A. et al. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 384, 2092–2101 (2021).
6.Muir, K.-L., Kallam, A., Koepsell, S. A. & Gundabolu, K. Thrombotic Thrombocytopenia after Ad26.COV2.S Vaccination. N. Engl. J. Med. 384, 1964–1965 (2021).
7.Planas, D. et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature (2021) doi:10.1038/s41586-021-03777-9.
8.Puranik, A. et al. Comparison of two highly-effective mRNA vaccines for COVID-19 during periods of Alpha and Delta variant prevalence. http://medrxiv.org/lookup/doi/10.1101/2021.08.06.21261707 (2021) doi:10.1101/2021.08.06.21261707.
9.Brown, C. M. et al. Outbreak of SARS-CoV-2 Infections, Including COVID-19 Vaccine Breakthrough Infections, Associated with Large Public Gatherings — Barnstable County, Massachusetts, July 2021. MMWR Morb. Mortal. Wkly. Rep. 70, (2021).
10.Werbel, W. A. et al. Safety and Immunogenicity of a Third Dose of SARS-CoV-2 Vaccine in Solid Organ Transplant Recipients: A Case Series. Ann. Intern. Med. L21-0282 (2021) doi:10.7326/L21-0282.
11.Venter, M. & Richter, K. Towards effective diagnostic assays for COVID-19: a review. J. Clin. Pathol. 73, 370–377 (2020).
12.Weissleder, R., Lee, H., Ko, J. & Pittet, M. J. COVID-19 diagnostics in context. Sci. Transl. Med. 12, eabc1931 (2020).
13.Horndler, L. et al. Flow cytometry multiplexed method for the detection of neutralizing human antibodies to the native SARS-CoV-2 spike protein. EMBO Mol. Med. 13, e13549 (2021).
14.Kubo, M. & Miyauchi, K. Breadth of Antibody Responses during Influenza Virus Infection and Vaccination. Trends Immunol. 41, 394–405 (2020).
15.van Schooten, J. & van Gils, M. J. HIV-1 immunogens and strategies to drive antibody responses towards neutralization breadth. Retrovirology 15, 74 (2018).
16.Tea, F. et al. SARS-CoV-2 neutralizing antibodies: Longevity, breadth, and evasion by emerging viral variants. PLOS Med. 18, e1003656 (2021).
17.Irani, V. et al. Molecular properties of human IgG subclasses and their implications for designing therapeutic monoclonal antibodies against infectious diseases. Mol. Immunol. 67, 171–182 (2015).
18.Martinez-Martin, N. et al. Cooperativity between T cell receptor complexes revealed by conformational mutants of CD3epsilon. Sci Signal 2, ra43. (2009).
Info/History
ARTICLE INFORMATION
https://www.medrxiv.org/content/10.1101/2021.08.12.21261952v1
doi https://doi.org/10.1101/2021.08.12.21261952
History August 14, 2021.
Copyright The copyright holder for this preprint is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission.
AUTHOR INFORMATION
Lydia Horndler1, Pilar Delgado1, Salvador Romero-Pinedo2, Marina Quesada2, Ivaylo Balabanov1, Rocío Laguna-Goya3, Patricia Almendro-Vázquez3, Miguel A. Llamas4, Manuel Fresno1, Estela Paz-Artal35, Hisse M. van Santen1, Stela Álvarez2, Asunción Olmo2 and Balbino Alarcón1*
1Centro de Biología Molecular Severo Ochoa, Consejo Superior de Investigaciones Científicas (CSIC), Universidad Autónoma de Madrid, 1 Nicolás Cabrera, 28049 Madrid, Spain
2VITRO SA, Avenida del Conocimiento 100, PT de la Salud, 18016 Granada, Spain
3Instituto de Investigación Sanitaria Hospital 12 de Octubre (imas12), 28041 Madrid, Spain
4EMPIREO Diagnóstico Molecular SL, 127 Fuencarral Street, 28004 Madrid, Spain
5Department of Immunology, Ophthalmology and ENT, Universidad Complutense de Madrid, Madrid, Spain
↵*Correspondence should be addressed to B.A. (balarcon@cbm.csic.es), Centro de Biología Molecular, Nicolás Cabrera 1, Universidad Autónoma de Madrid, Cantoblanco, Madrid 28049, Spain. Phone: 34-911964555; FAX: 34-911964420
